⚠️ Warning!
This site is archived, and therefore very out of date! See hrryt.github.io instead.
Harry Thompson
Check out my R package scenesetr!
Second-year undergraduate studying Natural Sciences at University of Cambridge. I'm interested in all things mathematical and computational in biology.
Experience with R
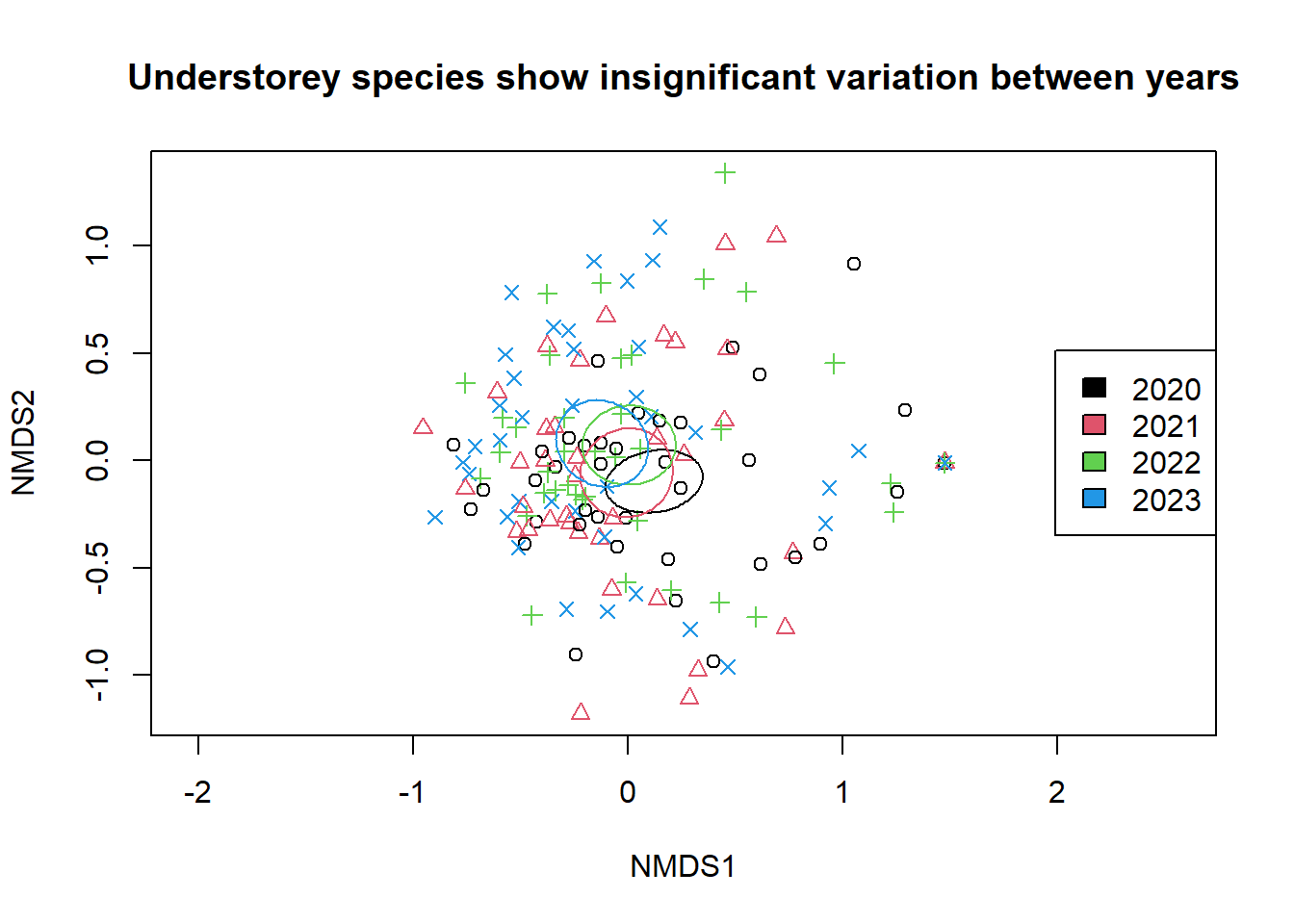
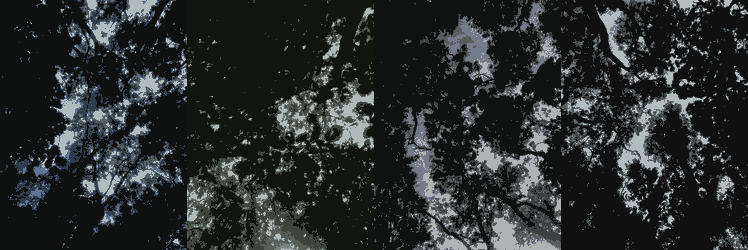
I've been using R since 2020 for a research project on the effects of ash dieback on the understorey of Haley Wood. R has so far provided me with the tools for ordination, image quantisation, exploratory data analysis and more. This project will continue through the years as we collect more data and learn more about the effects of plant pathology on ecosystem composition.
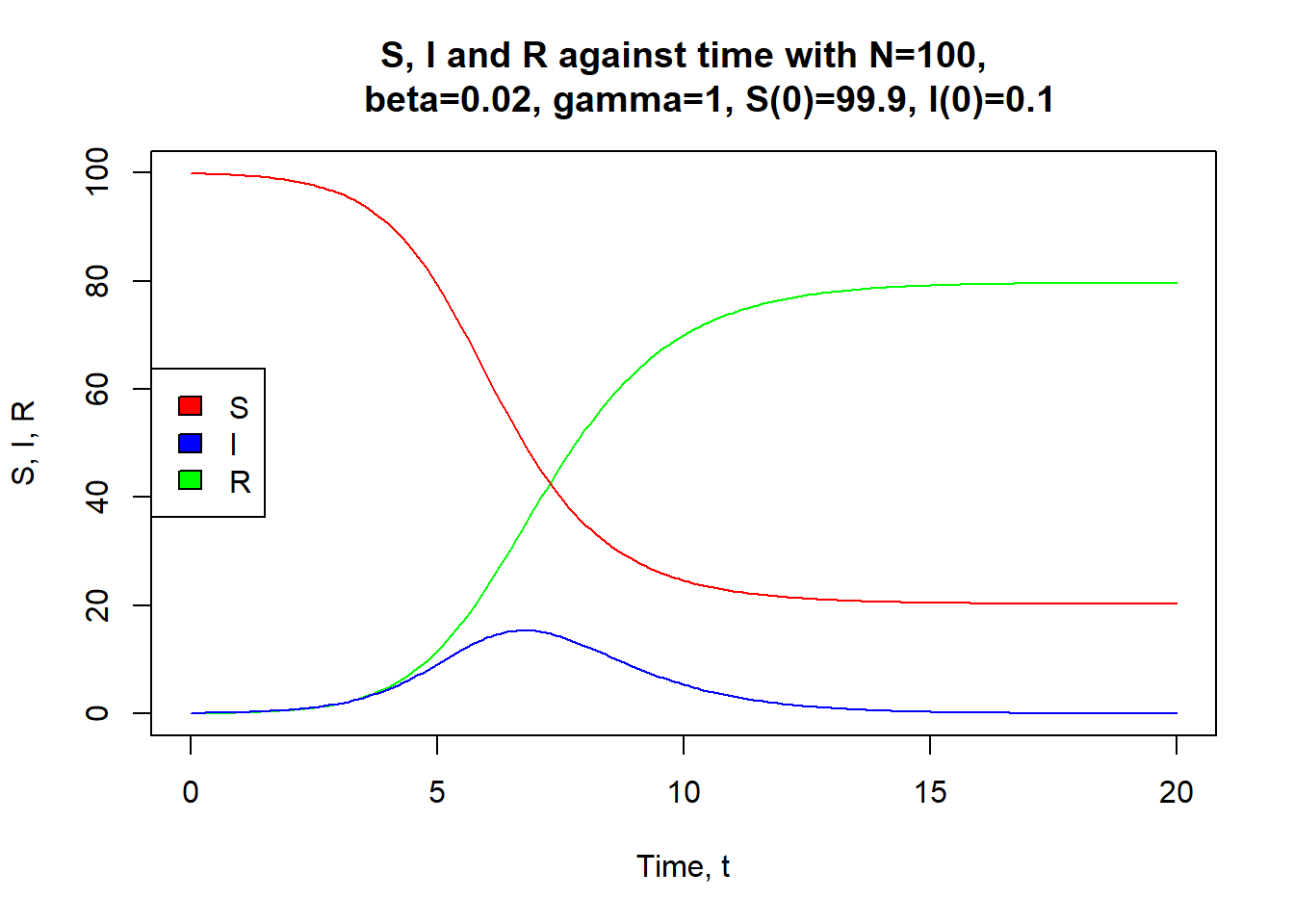
Through my degree, I've used R for epidemiological SIR modelling, dynamic programming for sequence alignment, generalised linear models, bootstrapping, spatial point pattern modelling and more.
I've also tutored a first-year Psychology student in the use of linear models and the tidyverse.
I've had fun wrangling and visualising data for a policy paper on atmospheric pollution. This has given me yet more experience with data analysis using the tidyverse and data.table, and has given an insight into accessibility not only for readers without a scientific background, but also for colourblind and visually impaired readers.
R Package Development
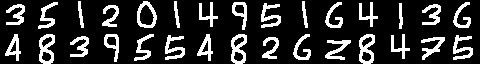
Recently, I've been working on a package to create neural networks in R. I decided I would challenge myself not to use any external packages, using pure linear algebra to fit sequential networks to data. So far, I've used this package to identify handwritten digits with an >60% success rate after one minute of training. Developing this package has required efficient R code and knowledge of R6 classes.

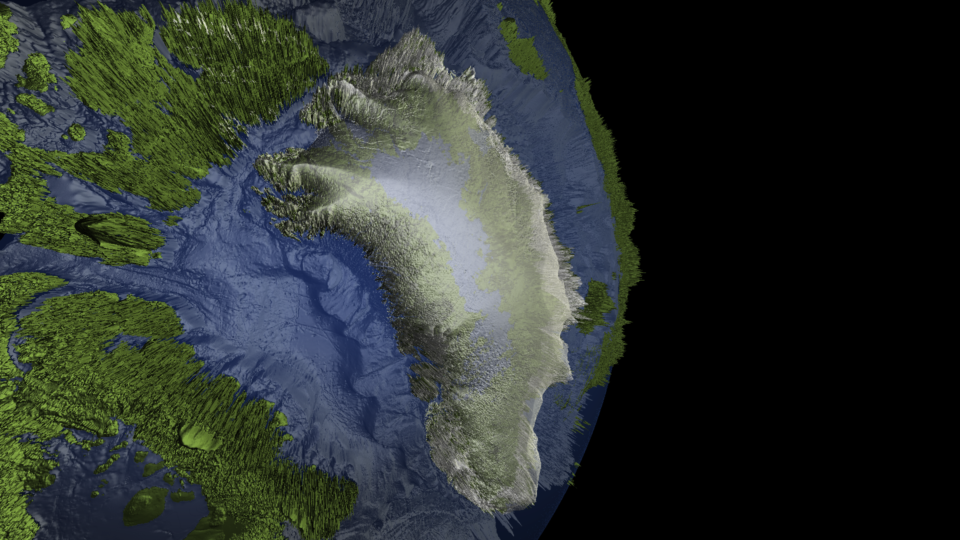
Where my knowledge of R has really developed has been in the development of a package for rendering 3-D scenes. In the making of this package, I've picked up a variety of skills, including:
- object oriented programming in S3, S4 and C++,
- profiling and optimisation of R code,
- problem solving with quaternions and matrices,
- integration of C++ code into R packages,
- dynamic linking of external C++ libraries in R packages,
- writing C++ code using OpenGL,
- working with complex data structures,
- collaborating with people who don't know R,
- handling geospatial contour and raster data,
- documentation and user-first programming,
- open-source publication on GitHub.
The above image of Greenland was generated using base R graphics, but I'm working on integrating OpenGL for boosted performance and graphics.

Experience with Python
As part of both Quantitative Environmental Science and Mathematical and Computational Biology this year, I've learned to use Python for a wide range of data analysis techniques, including:
- using a generalised linear model to successfully predict the 2022 World Cup,
- modelling the infection patterns of Salmonella bacteria between host phagocytes,
- analysing genetic data, including sequence alignment and Hidden Markov Model generation,
- training neural networks, performing K-means clustering, PCA and MDS,
- manipulating and analysing biological network data,
- solving differential equations and epidemiological models.
Studies
Before reading in Cambridge, I achieved A*A*A*A* in Maths, Biology, Chemistry and German at A-Level following eleven 9s and one 8 at GCSE. I also achieved an A in the Free Standing Mathematics Qualification.
First Year
- Mathematical Biology
- Biology of Cells
- Physiology of Organisms
- Evolution and Behaviour
Having achieved a First Class in Mathematical Biology and 2.1 elsewhere, the first year gave me invaluable experience over a broad range of biological disciplines.
Second Year
- Mathematical and Computational Biology (MCB)
- Quantitative Environmental Science (QES)
- Ecology, Evolution and Conservation (EEC)
The second year offers two new courses to Cambridge, MCB and QES, allowing me to explore further the role of maths and computation in biology and environmental science. EEC has been a great opportunity to develop my interest in global ecosystem functioning and evolutionary dynamics.
Looking to the Future
I aim to complete a Masters in Systems Biology or Quantitative Climate and Environmental Science. In the meantime, I'm spend the third year studying Genetics.